Using Gradient PCR to Determine the Optimum Annealing Temperature
By Vincent R. Prezioso and Axel Jahns, Eppendorf Scientific Inc.
Problems with the amplification of a specific DNA fragment using the PCR* are an everyday occurrence in the lab. Non-specific secondary bands may form after the PCR reaction, which hinder, or even prevent, further analysis (cycle sequencing, mutation detection, etc.) or an unequivocal assessment of the PCR result. In such cases, PCR conditions must be optimized. This is normally achieved by titrating the magnesium-, template-, primer-, dNTP- and Taq-Polymerase concentration, "Hot Start PCR," "Touch-down PCR," adding detergents, reducing the PCR cycles or by gradually increasing the annealing temperature. The selection of the annealing temperature is possibly the most critical component for optimizing the specificity of a PCR reaction. In most cases, this temperature must be empirically tested. The PCR is normally started at 5°C below the calculated temperature of the primer melting point (Tm).
However, the possible formation of non-specific secondary bands shows that the optimum temperature is often much higher than the calculated temperature (>12°C). This paper describes the use of the Gradient function of the Eppendorf Mastercycler Gradient to optimize a PCR reaction.
Gradient PCR is a technique that allows the empirical determination of an optimal annealing temperature using the least number of steps. This optimization can often be achieved in one experiment. The Eppendorf Mastercycler Gradient provides a gradient function that in one single run evaluates up to 12 different annealing, elongation, or denaturation temperatures. During the same run a number of possible concentration parameters can also be tested, row by row.
The goal of the experiment was to amplify a three hundred base pair fragment of the ribosomal spacer region of mycoplasma from H9 cell cultures. A previous experiment gave non-specific amplification at the calculated annealing temperature of 56.5°C (not shown).
Gradient PCR was used in the next experiment in order to determine the optimal annealing temperature. Using the gradient function of the universal block, a gradient of 53 to 67°C was set. This was achieved by setting a 14°C gradient and setting the annealing temperature at 60°C. The following test parameters were selected: denaturation 94,10 s, annealing 53°C -67°C, 15 s, elongation 72°C, 20 s, Taq-Polymerase 0.75 units. Figure 1 shows an agarose gel with the 12 samples that were loaded across the block. Clearly, the best conditions are found in well 10 (of the cycler) where the temperature was 63.5.
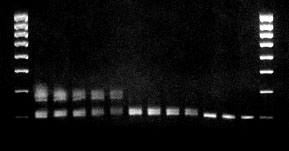
Figure 1: Experimental determination of optimal annealing temperature: The calculated primer annealing temperature was 56.5°C, the actual annealing temperature is 63.5°C. The ribosomal spacer region of mycoplasms from H9 cell cultures was amplified. Using the gradient function of the universal block, a gradient of 53 to 67°C was set. The following test parameters were selected: denaturation 10 s, annealing 15 s, elongation 20 s, Taq-Polymerase 0.75 units; duration of entire experiment: <1 h.
In Figure 2, the amplification shown in Figure 1 performed again, this time under optimized temperature conditions. In this experiment, the universal block was set to a uniform temperature in the annealing phase. The following test parameters were used: denaturation 94,10 s, annealing 63.5°C, 15 s, elongation 72°C, 20 s, Taq-Polymerase 0.75 units. Figure 2 shows the agarose gel with 12 samples loaded across the block.

Figure 2: Amplification shown in Figure 1 performed under optimized temperature conditions. In this experiment, the universal block was set to a uniform temperature in the annealing phase. The outstanding temperature homogeneity of the block ensures reproducible PCR results.
Gradient PCR may be used for multiple applications in PCR. For example, in the presence of 0.8 mM total dNTP concentration, a magnesium chloride titration series in 0.5 mM increments over the 1 mM to 4.5 mM range will determine the magnesium ion concentration, that produces the highest yield of a specific PCR product. In combination with a temperature gradient of 20°C across the block for the annealing step, important parameters of a specific PCR can be checked in just one run.
The gradient function of the Mastercycler is not restricted to the annealing temperature. The gradient function can also be used for optimization of the denaturation step, or the elongation temperature. DNA denaturation is a critical step in the PCR process and is often the step where PCR experiments fail. The temperature range for denaturation of most DNA samples is 94°C to 96°C. However, special target DNAs require a different denaturation temperature to achieve an optimal result. The gradient would determine the lowest possible denaturation temperature providing high yield of amplified DNA.
The exposure of the Taq Polymerase to unnecessary high temperatures would decrease the enzyme activity over the PCR run, which would have to be compensated by using more enzyme.
For determining the optimal denaturation temperature of these templates, the gradient function of the Mastercycler can be set from 90 - 99 °C. An initial denaturation step of 1 to 5 minutes (3 - 10) could further optimize the reaction. Harmful nucleases in the sample will be inactivated, ensuring the complete denaturation of complex templates such as genomic DNA.
In special cases the primer annealing temperature may need to be raised as high as the extension temperature. In fact, high-temperature annealing should result in enhanced specificity, because the hybridization of the primer to the template DNA occurs under more stringent conditions. Combining primer annealing and primer extension steps results in a two-step PCR protocol. Primer extension, in most applications, occurs effectively at a temperature of 72 °C and seldom needs optimization. In contrast, in two-temperature PCR experiments, the annealing-extension temperature may be in the range of 60 to 70 °C. Again, the optimal temperature can easily be determined with the gradient function of the Mastercycler.
For more information: Michelle Lozito, Marketing Coordinator, Eppendorf Scientific Inc., One Cantiague Rd., PO Box 1019, Westbury, NY 11590. Tel: 800-421-9988, ext. 375. Fax: 516-876-8599.
* The Polymerase Chain Reaction (PCR) is protected by patent. The patent is held by Hoffmann-La Roche.
Eppendorf Scientific, Inc., One Cantiague Road, Westbury, NY 11590. Tel: (516) 334-7500; Fax: (516) 334-7506.
