New Developments In mRNA Vaccine Efficacy Show Promise
By Olivier Laczka, Ph.D., chief scientific officer (autoimmunity and inflammation), Noxopharm
From both a public health and economic perspective, the COVID-19 vaccine rollout has been an enormous success. It also has given mRNA-related research added momentum, and the field is now rapidly growing as various organizations and companies increasingly invest in the potential of this technology to go beyond COVID-19 vaccines and address a wide range of diseases, including cancer. However, there are still challenges at the core of contemporary mRNA vaccines that need to be met before the technology can reach its full potential.
One of the key issues is vaccine-induced innate immune activation (reactogenicity), which leads to side effects like fever and headaches that are dose-limiting. The current strategy to resolve this involves incorporation of modified nucleosides — the most well-known of which is pseudouridine, first described by 2023 Nobel Prize winners Katalin Karikó and Drew Weissman in their influential 2005 Immunity paper — into mRNA during in vitro transcription. Underscoring the importance of mitigating mRNA reactogenicity for future vaccine development, CureVac’s unmodified mRNA COVID-19 vaccine was unsuccessful in Phase 3 clinical trials during the pandemic. However, as this article will discuss, significant caveats with the current base modification approach remain and need to be addressed.
Activation Of Innate Immunity
Following administration, lipid-nanoparticle conjugated mRNA vaccines are taken up by phagocytic cells at the injection site, where a proportion of the mRNA is translated to make the antigen that will weaponize the body’s adaptive immune system against the infection or malignancy. Unfortunately, some of this mRNA also triggers activation of the innate immune system by tripping a series of sensors that are designed to detect foreign, or non-self, nucleic acids. These include the endosomal receptors Toll-like receptor 7 (TLR7) and TLR8, which recognize degradation fragments of guanidine- and uridine-rich RNAs respectively, and the cytosolic receptors RIG-I and MDA-5, which recognize cytoplasmic double-stranded and 5′-triphosphate-modified RNA, both potential contaminants of T7-synthesized mRNAs.
Activation of innate immunity acts as a first line of defense against invading pathogens and results in inflammation that we feel as fever, myalgia, headache, and fatigue — all common symptoms of a cold or flu, and also common vaccine side effects.1 In the context of mRNA vaccines, the endosomal receptor TLR7 has been shown to play a dominant role in driving innate immune activation, termed reactogenicity,2 which has a number of implications for the design and manufacture of RNA technologies going forward.
The success of the existing mRNA vaccines was, in large part, made possible by the work of Katalin Karikó’s team in the mid-2000s, which showed that incorporation of modified nucleosides – many of which are naturally occurring in human RNA – into synthetic mRNA dampens their detection by nucleic acid sensors, including TLR7.3,4 Both Pfizer-BioNTech and Moderna used a modified N1-methylpseudouridine (m1ψ) in place of uridine in their COVID-19 mRNA vaccines, which enabled mRNA doses of up to 100 mg (in the case of Moderna’s mRNA-1273 vaccine) to be well tolerated by recipients. In contrast, CureVac pursued an unmodified mRNA vaccine against COVID-19 and found that the maximum tolerated dose of mRNA was only 12 mg, which failed to generate a protective adaptive immune response in more than 50% of patients, leading to an unsuccessful Phase 3 clinical trial.
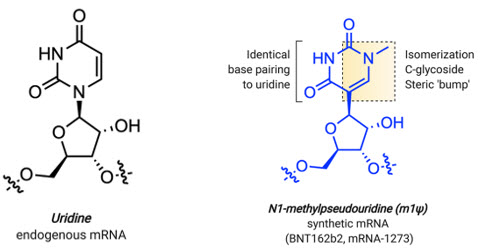
Figure 1: Structural features of uridine and m1Ψ. Source: “Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines.” Kellie D. Nance and Jordan L. Meier, ACS Central Science 2021 7 (5), 748-756. Use of the image is licensed under Creative Commons CC-BY-NC-ND 4.0.
Overcoming The Limitations Of Pseudouridine
Reactogenicity executes a bipartite hit on mRNA vaccine effectiveness. First, side effects of vaccinations are dose-limiting, even for modified mRNA, and also have a negative impact in terms of public perception, thereby reducing vaccine uptake in the population. Second, too much reactogenicity inhibits mRNA translation, resulting in a weaker adaptive immune response and reduced vaccine efficacy.
The most profound limitation of m1ψ and other base modifications as a strategy to mitigate mRNA reactogenicity by TLR7/8 is that its mechanism of action is not well characterized. Two possibilities exist: (1) that the substitution of m1ψ for uridine prevents the cleavage of mRNA into short immune-activating fragments by motif-selective endonucleases, such as RNase T2 or (2) that m1ψ impedes binding of mRNA degradation fragments to TLR7 and TLR8. Ultimately, without an in-depth understanding of how base modification, like m1ψ incorporation, works to limit reactogenicity, it is difficult to develop tools to accurately predict the reactogenicity of potential RNA sequences during their therapeutic development, meaning that more RNA technologies are likely to fail in clinical trials despite base modification. Additionally, base modification as a strategy to limit reactogenicity is incompatible with some emerging technologies, e.g., self-amplifying (sa)RNAs.
Interestingly, preliminary Phase 1 data released by CureVac in January 2023 showed that mRNA sequence identity greatly impacts the reactogenicity of even modified mRNA, with their m1ψ-modified Flu-SV-mRNA vaccine being significantly more reactogenic than their m1ψ-modified CV0501 COVID-19 vaccine at the same doses.5 These results further highlight the unpredictability and lack of control inherent in the use of pseudouridine, with the variability in percentage of pseudouridine bases incorporated, as well as the positioning of modified bases, within an mRNA sequence having an unpredictable impact on innate immune activation. It has long been understood that a small dose of innate immune activation is desirable for a strong adaptive immune response — a phenomenon called the adjuvant effect that has been exploited in the co-delivery of TLR agonists with many vaccines. With new research by Drew Weissmann and colleagues now suggesting that pseudouridine’s wholesale abrogation of innate immune signalling may actually impede robust adaptive immunity to mRNA vaccines,6 better characterized and more tuneable alternatives are urgently needed for the field to progress.
Novel 3-Base Oligonucleotides
Since the Karikó and Weissman breakthrough Immunity paper in 2005, significant work has been undertaken to understand how innate immune sensors, like TLR7 and TLR8, function to detect nucleic acids. Crystal structures for both RNA-bound receptors, published in 2015 for TLR8 and 2016 for TLR7, have now revealed that each receptor has two distinct binding sites, one for guanosine or uridine and one for a short RNA fragment.7,8 It is now proposed that endocytosed RNA is sequentially degraded into short RNA fragments (>2 bases) by select endonucleases, including RNase T2,9 or processed into single nucleosides by exonucleases, such as guanosine and uridine. Guanosine and uridine can then bind to site 1 of TLR7 or TLR8, respectively, while RNA degradation intermediates of a few nucleotides bind to site 2. Binding of guanosine or uridine to site 1 then synergizes with engagement of site 2 by the short degradation intermediates to promote downstream inflammatory signalling and subsequent reactogenicity.
More recently, Associate Professor Michael Gantier of the Hudson Institute of Medical Research in Melbourne, Australia, has been working to define how chemically modified synthetic RNA fragments can be used to modulate signalling via TLR7 and TLR8.10 This research, which underpins our Sofra technology platform at Noxopharm, has revealed that sequences as short as 3-bases can govern TLR7 and TLR8 sensing, acting as “immune codons” that help to delineate self from non-self RNAs. Critically, the capacity to rationally design and synthesize such chemically modified 3-base oligonucleotides to modulate TLR7 and TLR8 sensing offers a novel approach to control mRNA reactogenicity driven by these innate immune sensors. Indeed, these short oligonucleotides represent a new class of immunomodulatory therapeutics.
Building on this new modality to control TLR7/8 sensing of RNA, our approach at Noxopharm proposes to co-deliver short oligonucleotides, termed SOF-VAC, with any therapeutic mRNA to mitigate TLR7/8 sensing. This strategy has a number of key advantages over base modification of mRNA itself. First, the molecular mechanism of action of the 3-base oligonucleotides is well defined and independent of mRNA sequence identity, decreasing the risk of unintended off-target reactogenicity when using novel sequences. Second, this approach opens up opportunities for fine-tuning the level of reactogenicity, allowing for enhanced adaptive immune responses. Third, while base modification of mRNA reduces ex vivo transcriptional fidelity and can decrease T7 polymerase synthesis yields by as much as 50 %in some cases,11 co-delivery of inhibitory oligonucleotides with mRNA has no impact on mRNA yields ex vivo. Fourth, based on our latest in vivo studies, we anticipate that the inhibitory activity of 3-base oligonucleotides on TLR7/8 will allow for higher mRNA doses to be delivered before dose-limiting reactogenicity is reached, thereby facilitating the development of multivalent vaccines.
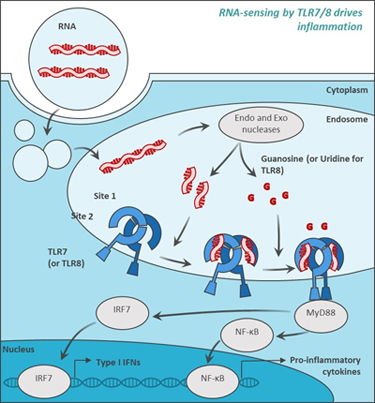
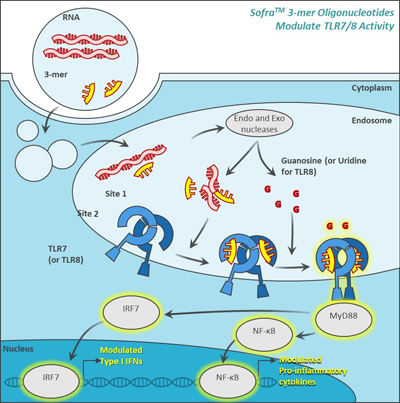
Figure 2: Novel ultra-short oligonucleotides potently potentiate or inhibit TLR7/8 activity in a sequence-dependent manner. Endocytosed RNA is degraded into single nucleosides, including guanosine, uridine, and degradation intermediates of several bases. Guanosine and uridine bind to site 1 of TLR7/8 to induce TLR7 and TLR8 activation, respectively. Degradation intermediates bind to site 2 of TLR7/8 and potentiate sensing. Chemically modified 3-mer oligonucleotides interfere with site 1 activation through direct binding to TLR7/8.
Beyond COVID-19 Vaccines
The potential to develop a new class of immunomodulatory therapeutics, while improving efficiencies in the manufacturing of mRNA vaccines, could lead to significant progress in the ongoing mRNA story, which many commentators believe is still in its relative infancy.
The positive outcomes brought about by the COVID-19 vaccines are the clearest indication to date of the ability of this technology to be rapidly adapted in the face of severe and urgent public health needs, as well as its potential to play a leading role in meeting the challenges posed by a myriad of other diseases. Chief among these is cancer, and international research efforts are now ramping up around the world as governments and industry participants devote increasing resources and investments to preclinical and clinical studies in this space. We believe these efforts have a high chance of success in the medium to long term, especially as our collective understanding of mRNA technologies deepens and emerging technologies, such as saRNAs and circular RNAs, play a greater role in R&D programs.
Even more encouraging is a growing awareness of the potential of mRNA technology among the public, where attention is now clearly beginning to focus on the next steps in the wake of the pandemic. Such understanding will help build support and funding for future developments, leading to a greater opportunity to educate populations about a variety of new RNA-based therapeutics as, and when, they are rolled out in the years to come.
References
- Park J.W., et al. mRNA vaccines for COVID-19: what, why and how. 2021. International Journal of Biological Sciences, 17(6): 1446-1460.
- Verbeke R., et al. Innate immune mechanisms of mRNA vaccines. 2022. Immunity, 55(11): 1993-2005.
- Karikó K., et al. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. 2005. Immunity, 23(2): 165-175.
- Karikó K., et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. 2008. Molecular Therapy, 16(11): 1833-1840.
- CureVac. Preliminary Phase 1 Data from Joint COVID-19 and Flu mRNA Vaccine Development Programs. January 2023 presentation. https://www.curevac.com/wp-content/uploads/2023/01/20230106-Presentation_CureVac-Announces-Positive-Data-on-Joint-COVID-19-and-Flu-mRNA-Vaccine-Development-Programs.pdf
- Han, X., et al. Adjuvant lipidoid-substituted lipid nanoparticles augment the immunogenicity of SARS-CoV-2 mRNA vaccines. 2023. Nature Nanotechnology, 18:1105-1114.
- Tanji H., et al. Toll-like receptor 8 senses degradation products of single-stranded RNA. 2015. Nature Structural Molecular Biology, 22(2): 109-115.
- Zhang Z., et al. Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. 2016. Immunity, 45(4): 737-748.
- Greulich W., et al. TLR8 Is a Sensor of RNase T2 Degradation Products. 2019. Cell, 179(6): 1264-1275.
- Alharbi A.S., et al. Rational design of antisense oligonucleotides modulating the activity of TLR7/8 agonists. 2020. Nucleic Acids Research, 48(13): 7052-7065.
- Kauffman K.J., et al. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. 2016. Biomaterials. 109: 78-87.
About The Author:
 Olivier Laczka is chief scientific officer (autoimmunity and inflammation) at Noxopharm. An experienced leader in drug development, biopharmaceuticals, biotechnology, and diagnostics, his skillset includes immunoassays, patents, scientific writing and grants, and intellectual property and research funding, along with managing effective collaborations with commercial organizations, academic institutions, governments, and private entities at the international level. Prior to joining the company in 2018, Laczka was a senior research scientist with 3M, PriME Biologics, and AEGROS Innovations. He also held roles in academic institutions such as UTS Sydney, Macquarie University, and the Institute of Microelectronics of Barcelona.
Olivier Laczka is chief scientific officer (autoimmunity and inflammation) at Noxopharm. An experienced leader in drug development, biopharmaceuticals, biotechnology, and diagnostics, his skillset includes immunoassays, patents, scientific writing and grants, and intellectual property and research funding, along with managing effective collaborations with commercial organizations, academic institutions, governments, and private entities at the international level. Prior to joining the company in 2018, Laczka was a senior research scientist with 3M, PriME Biologics, and AEGROS Innovations. He also held roles in academic institutions such as UTS Sydney, Macquarie University, and the Institute of Microelectronics of Barcelona.
