Application Note: 96 Well Streptavidin-Coated Microplates

 Streptavidin microplates are coated with streptavidin, a 60 KD protein isolated from Streptomyces avidinii. The purified protein is bound to the wells of polystyrene microplates via a proprietary coating technology. This coating technology ensures:
Streptavidin microplates are coated with streptavidin, a 60 KD protein isolated from Streptomyces avidinii. The purified protein is bound to the wells of polystyrene microplates via a proprietary coating technology. This coating technology ensures:
- high binding capacity of biotin
- high coating homogeneity
- low leaching of streptavidin (<6 ng per well)
- high resistance to commonly used detergents
In addition, streptavidin-coated microwell plates are pre-blocked for immediate use. High capacity binding assays of biotinylated single and double stranded DNA, peptides, proteins, and small organic molecules can be performed on streptavidin-coated microplates.
Reaction Volume
Streptavidin is coated at a reaction volume of 100 /well. The wells are blocked at 200 µL/well.
Binding Capacity
In saturation and competitive binding assays performed on this product, binding of 325 pmoles of d-biotin per well is observed. The binding capacity of larger molecules labeled with biotin will possibly be less than that of biotin via steric factors associated with the specific molecule.
Storage and Handling Conditions
For optimal performance, the unopened product should be stored in a dry place at 2–30°C. Under these storage conditions, the product is stable for two years. Once opened, it is suggested that the product be used immediately.
Not recommended for assays at >60°C because of potential plate instabilities.
Sample Protocols for the Use of Streptavidin-Coated Microplates
Plate Viability Assay
To validate the viability of the streptavidin surface follow the procedure below:
- Using Sigma catalog item P 9568 (Biotinylated HRP), dissolve 1 mg in 1 ml of PBS + 0.05% Tween20 (P 3563). Dilute the 1 mg/ml stock 1:50,000–1:100,000 in PBS + 0.05% Tween-20 and add 100 µL per well. As a negative control, add 100 µL of a similar dilution of streptavidin-peroxidase (S 5512) to a separate set of wells.
- At room temperature, incubate the wells for 30 minutes.
- Wash the wells three times, 300 µL per well, with PBS + 0.05% Tween-20.
- After discarding the final wash, add 100 µL per well of TMB substrate (T 8665).
- Incubate the wells for 15 minutes before reading in a microplate absorbance reader or spectrophotometer. If desired, the reaction may be stopped with the addition of 0.5M H2SO4 (50 per well). An absorbance of at least 1.5 will be observed at 655 nm for a non-stopped reaction or 450 nm for a stopped reaction.
Peptide and Protein Binding
- Prepare a solution of the biotinylated protein or peptide in 0.05M bicarbonate buffer pH 9.6, PBS pH 7.4 or TBS pH 7.5. A starting concentration of 1-10 µg/ml should be used if the optimal concentration is not known.
- Add up to 100 µL of the solution per well and allow the samples to incubate for 1-2 hours at a temperature range of 18 to 30°C.
- Wash the wells three times, 300 µL per well, with PBS or TBS + 0.05% Tween-20.
- Incubate the wells with 100 µL of an appropriately diluted primary antibody in PBS or TBS + 0.05% Tween-20 for 30-60 minutes.
- Wash the wells three times, 300 µL per well, with PBS or TBS + 0.05% Tween-20.
- Incubate the wells with 100 µL of an appropriately diluted enzyme labeled secondary antibody in PBS or TBS + 0.05% Tween-20 for 30-60 minutes.
- Wash the wells three times, 300 µL per well, with PBS or TBS + 0.05% Tween-20.
- After addition of an appropriate substrate, the wells
are ready for detection in various modes (colorimetry, chemiluminescence, or fluorescence).
PCR Products
- Prepare the biotinylated PCR product for addition onto streptavidin-coated microplates by diluting the reaction 1:10-1:50 in PBS + 0.05% Tween-20.
- Apply 100 µL per well of the diluted product and allow the sample to incubate for 30-60 minutes at 25-37°C.
- Bound PCR products are denatured by adding 100 µL per well of 0.5M NaOH. Incubate for 5-10 minutes.
- Wash the wells three times, 300 µL per well, with PBS + 0.05% Tween-20 to remove the nonbiotinylated, complimentary strand of the PCR product.
- Add 200 µL per well of a hapten labeled oligonucleotide which is complimentary to the biotinylated strand. Use 0.05–0.5 pmole of labeled oligonucleotide per well. Hybridize in the presence of 5X SSC, 0.3% Tween-20,
1% BSA. Allow the hybridization to proceed for 30-60 minutes at 37-50°C.
- Wash the wells three times, 300 µL per well, with PBS + 0.05% Tween-20.
- Add 100 µL per well of an appropriately diluted detection conjugate in PBS + 0.05% Tween20. Incubate for 30–60 minutes at RT.
- Wash the wells five to six times, 300 µL per well, with PBS + 0.05% Tween-20.
- Proceed to the use of the specific substrate for detection.
Detection
A number of alternatives exist for detecting molecules labeled with common haptens. Below is a table of indirect detection systems for two haptens, namely biotin and fluorescein.
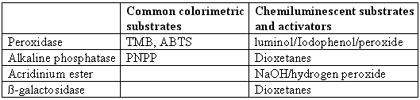
Optimization of Elisa Results
There are four major areas where detection results can be optimized: nonspecific binding, antibody affinity, wash conditions, and conjugate concentration.
- Nonspecific binding: Factors that contribute to nonspecific binding are ionic interactions, hydrophobic interactions, and cross-reactivity. To reduce nonspecific binding, changes in conjugate concentrations and wash buffers can be made. Users are encouraged to modify buffers with components in the ranges of the various conditions listed below.
![]()
- Wash Conditions: To limit reversible nonspecific binding interactions, at least three wash steps are recommended.
- Antibodies and Conjugates: For optimal signal performance, the user is encouraged to
use high affinity antibodies and conjugates. Commercially obtained antibodies and conjugates should be used at the concentrations suggested by the supplier.
Recommended Reagents Available Through Sigma Aldrich Corp.

1-800-325-3010
General References
- Wilchek M, Bayer EA eds: Meth Enzymol, Avidin-Biotin Technology, 184, 1990.
- Crowther JR ed: "ELISA: Theory and Practice" Methods in Molecular Biology, Vol 42 (Humana Press, Totowa, NJ, 1995), Catalog # Z36,415-0.
- Dekonenko A, Ibrahim MS, Schmaljohn CS: A Colorimetric PCR-Enzyme Immunoassay to Identify Hantaviruses. Clin and Diagnostic Virology 8:113-121, 1997.
- Fujita S et al.: Microtitration Plate Enzyme Immunoassay To Detect PCR-Amplified DNA from Candida Species in Blood. J Clin Microbiol 33:962-967, 1995.
- Guenthner PC, Hart CE: Quantitative, Competitive PCR Assay for HIV-1 Using a Microplate-Based Detection System. Biotechniques 24:810816, 1998.
- Rong R et al., Two-Site Immunofluorometric Assay of Intact Salmon Calcitonin with Improved Sensitivity. Clin Chem 43:71-75, 1997.
- Vesanen M: et al.: Detection of Herpes Simplex Virus DNA in Cerebrospinal Fluid Samples Using the Polymerase Chain Reaction and Microplate Hybridization. J Virol Meth 59:1-11, 1996.
- Benotmane AM et al.: Nonisotopic Quantitative Analysis of Protein-DNA Interactions at Equilibrium. Anal Biochem 250:181-185, 1997.
- Verhaegen M, Christopoulos TK: Quantitative Polymerase
Chain Reaction Based on a Dual-Analyte Chemiluminescence Hybridization Assay for Target DNA and Internal Standard. Anal Chem 70:4120-4125, 1998.
For Research Use Only. Not for use in diagnostic procedures.
For more information: Christine Pennachio, Market Development Coordinator, Corning. Tel: 978-635-2368. Fax: 978-635-2490.

