Structural Bioinformatics, Quest Launch HIV Drug Pharmacogenomic Databases

Each QSVdBase database module contains a large collection of high-quality protein structures generated from the genetic sequence variants (polymorphisms) of a single drug target derived from many individuals. These structure data provide a definitive key to understanding differences in interactions between a drug or potential drug and the structural variations of its intended target, which is critical to optimal drug design and the selection of the optimal drug candidate for initiation of human trials.
QSVdBase converts sequence information into statistically-relevant three-dimensional structural information. The HIV-RT and HIV-PR modules contain more than 15,000 unique structures (in aggregate) derived from individual patient samples, along with drug resistance data and structures for all currently approved drugs docked into the active site.
The database interface provides query tools and is SQL-compatible for more complex queries. Large sample size provides a representative spectrum of HIV mutations for meaningful statistics and provides trend data for changes in mutational frequencies over a period of two years. Sampling is current and on-going with the addition of approximately 1,000 sequences/structures per month. The sequence data is generated by Quest Diagnostics' CAP and ISO9001 certified procedures.
HIV drug resistance is a critical factor when developing new AIDS drugs. SBI believes the information in QSVdBase will allow pharmaceutical companies to make better choices of candidate compounds before tens of millions of dollars are spent on clinical trials.
"Researchers and physicians may be familiar with so-called mutation lookup tables, which list amino acid positions and known mutations which correlate with resistance," said Bernard L. Kasten, Quest Diagnostics' chief medical officer. "QSVdBase is deeper, richer, and more accessible than public HIV mutation tables and adds structural, temporal and patient-specific information that is not available elsewhere."
Which Drugs Will Be Resistant?
The database was developed through Quest's extensive experience in mapping out HIV mutations using their HIV phenotype test. By analyzing the HIV genomes of thousands patients failing reverse transcriptase or HIV protease treatment, Quest has generated a huge raw database of HIV sequences, which SBI has converted into the new database products.
Both QSVdBases should become indispensible for designing drugs based on either reverse transcriptase or protease inhibition, says SBI vice president Dean Goddette. "Preliminary data on new HIV drugs coming out during the next few years suggest that some HIV strains are already resistant to these agents. If you're a drug company you don't want to find that out after a drug is coming onto the market. You want to know early in the discovery process to try to design drugs that target the drug-resistant variants as well as the wild type HIV virus."
Goddette also sees the new databases as a tool for ranking new drug candidates preclinically. "The databases can also generate temporal information, so you can follow patients over time," Goddette told DrugDiscovery Online. "And ultimately, what we want to work with quest on is to design new clinical diagnostic tests based on knowledge of the sequence and 3D structure, so we can essentially have a computational phenotype test.
"Today, drug companies have access to tables of mutations known to cause resistance, but this information is static and based on very small numbers of samples. Plus there's no simple test that can be done, when a patient comes into the doctor's office, to measure directly if their HIV is resistant or nonresistant. In the long run, we're shooting for a computational phenotype so that from the sequence we can accurately predict if a virus is resistant or not."
About SBI
Privately-held Structural Bioinformatics specializes in computational proteomics—the large-scale generation and use of protein structure and protein structural properties. SI has developed advanced computational technologies used to generate highly refined three-dimensional structural models of proteins from primary gene sequence data, which compare well to crystal structures. SI's products allow discovery-stage companies to perform primary drug-discovery screening in silico on millions of compounds present in SBI's scaffold-specific virtual combinatorial libraries (CombiLib modules) or customers' own in-house compound collections.
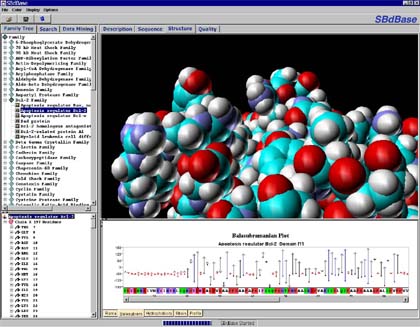
Screen shot of the Structural Bioinformatics SBdBase version 2, a database of modeled proteins structures, showing the family list and a CPK model of the binding pocket of Bcl-2, a protein involved in apoptosis (programmed cell death). Overexpression of Bcl-2 is implicated in confering immortality to certain types of cancer cells.
SBI offers two subscription database products. The first, SBdBase, composed of thousands of structural models of proteins from more than 150 distinct families, can accelerate lead discovery and optimization while enabling rational experimental design across multiple disciplines. The second product, SVdBase, is a growing series of database modules each containing structural models generated from genetic variations in a single, medically important gene. SBI is big on collaborative R&D. Current partners include BioChem Pharma Inc., Yamanouchi Pharmaceutical Co. Ltd. and DuPont Pharmaceuticals Co.
For more information: Dean Goddette, VP of Marketing, Structural Bioinformatics, Inc., 10929 Technology Place, San Diego, CA 92127. Tel: 858-675-2400. Fax: 858-451-3828
By Angelo DePalma
